Welcome to menoci.io
menoci is a collection of open source Drupal extension modules that can be used to rapidly deploy an integrated website and data management portal for research projects in the biomedical and life sciences.
The menoci modules are actively developed and maintained by the University Medical Center Göttingen, Department of Medical Informatics, and Scientific Data Center Göttingen, GWDG.
Architecture
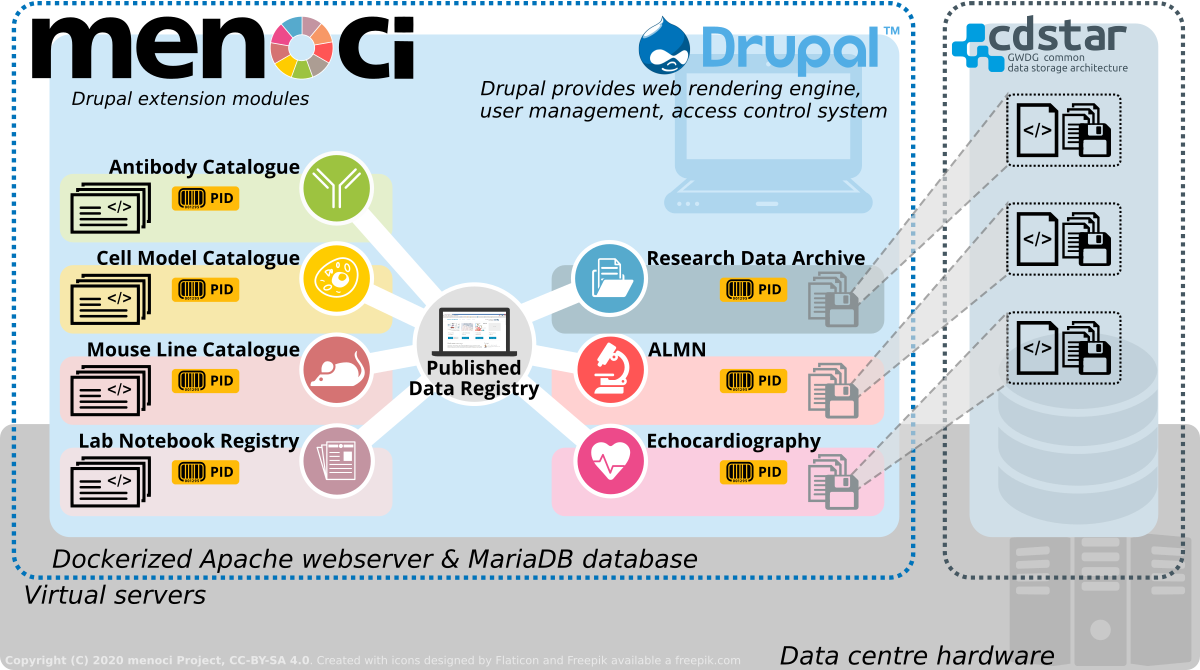
Based on the Drupal web content management system, menoci introduces multiple extension modules that support scientific data management for life science research projects.
Features
Features of menoci include:
- Manage and display the scientific track record of your project in a standardized format (peer-reviewed article, published datasets, preprints, etc.)
- Document and display all resources linked to the published data
- Keep track of key assets for your biomedical experiments:
- Antibodies,
- Mouse lines and mice,
- Cell lines* and cell models (iPSC, hiPSC)
- Laboratory notebooks*
- Store your research data in CDSTAR and publish datasets with persistent resolvable identifiers from the ePIC consortium.
*) The menoci modules "Cell Model Catalogue" and "Lab Notebook Registry" are not currently included in the default software distribution but are freely available upon request.
Reference projects
| Project | Description | Duration |
|---|---|---|
| CRC 1002 | The project investigates heart insufficiency in translational medicine. Origin of the menoci software. | 2012 - today |
| CRC 1190 | Compartmental Gates and Contact Sites in Cells. more information | 2014 - today |
| CRC 1286 | The project is specialised in Quantitative Synaptology and promotes a modern infrastructure including super-resolution imaging facilities. more information | 2017 - today |
| CURE-PLaN | Cure PhosphoLambaN induced cardiomyopathy more information | 2019 - 2023 |
| MBExC | Multiscale Bioimaging (MBExC) is a Cluster of Excellence of the University of Göttingen more information | 2020 - today |
| TRR 274 | Checkpoints of Central Nervous System Recovery more information | 2020 - today |
| FOR 2705 | Dissection of a Brain Circuit: Structure, Plasticity and Behavioral Function of the Drosophila Mushroom Body more information | 2020 - today |
| FOR 2848 | Nanoscale Architecture and Hereogeneity of the Mitochiondrial Inner Membrane more information | 2022 - today |
| CRC 1565 | The project is specialised in Molecular mechanisms and interplay of gene expression processes. more information | 2023 - today |
| EKFZ | Else Kröner Fresenius Center for Optogenetic Therapies (EKFZ) more information | 2025 - today |
| CRC 1690 | Disease Mechanisms and Functional Restoration of Sensory and Motor Systems. more information | 2025 - today |
Literature
The menoci software project has been covered in the following scientific articles:
- Freckmann, Luca, Henke, Christian, Kossen, Robert, Weber, Linus, Sax, Ulrich, Nussbeck, Sara Y., Kusch, Harald. 2023. Approaching automation of multiple instance orchestration of the menoci web portal. Bausteine Forschungsdatenmanagement. doi:10.17192/bfdm.2023.5.8608 . View in menoci
- Kusch H, Kossen R, Suhr M, Freckmann L, Weber L, Henke C, Lehmann C, Rheinländer S, Aschenbrandt G, Kühlborn LK, Marzec B, Menzel J, Schwappach B, Zelarayan LC, Cyganek L, Antonios G, Kohl T, Lehnart SE, Zoremba M, Sax U, Nussbeck SY. Management of Metadata Types in Basic Cardiological Research. Stud Health Technol Inform 2021 doi:10.3233/shti210542 . View in menoci
- Suhr M, Lehmann C, Bauer CR, Bender T, Knopp C, Freckmann L, Öst Hansen B, Henke C, Aschenbrandt G, Kühlborn LK, Rheinländer S, Weber L, Marzec B, Hellkamp M, Wieder P, Sax U, Kusch H, Nussbeck SY. Menoci: lightweight extensible web portal enhancing data management for biomedical research projects. BMC Bioinformatics 2020 doi:10.1186/s12859-020-03928-1 . View in menoci
- Suhr M, Lehmann C, Bauer CR, Bender T, Knopp C, Freckmann L, et al. menoci: Lightweight Extensible Web Portal enabling FAIR Data Management for Biomedical Research Projects. Feb 2020, doi:10.1186/s12859-020-03928-1View in menoci
- Lehmann C, Suhr M, Umbach N, Cyganek L, Kleinsorge M, Nussbeck SY, et al. Leaving spreadsheets behind – FAIR documentation and representation of human stem cell lines in the Collaborative Research Centre 1002. GMDS 2019, Dortmund. doi:10.3205/19gmds037 . View in menoci
- Rheinländer S, Aschenbrandt G, Nussbeck SY, Suhr M, Kusch H. Towards standardized documentation of mouse lines in biomedical basic research. GMDS 2019, Dortmund. doi:10.3205/19gmds038 . View in menoci
- Suhr M, Jahn N, Mietchen D, Kusch H. Wikidata as semantic representation platform of the scientific achievements of the biomedical Collaborative Research Centre 1002. GMDS 2018, Osnabrück. doi:10.3205/18gmds173 . View in menoci
- Kusch H, Schmitt O, Marzec B, Nussbeck SY. Data organization of a clinical Collaborative Research Center in an integrated, long-term accessible Research Data Platform. GMDS 2015, Krefeld. doi:10.3205/15gmds104 . View in menoci
Contact
This project is currently maintained by
- Linus Weber, linus.weber@med.uni-goettingen.de, University Medical Center Göttingen, Department of Medical Informatics
- Christian Henke, christian.henke@med.uni-goettingen.de, University Medical Center Göttingen, Department of Medical Informatics
- Harald Kusch, harald.kusch@med.uni-goettingen.de, University Medical Center Göttingen, Department of Medical Informatics
Introduction
menoci is a collection of Drupal extension modules that can be used to rapidly deploy an integrated website and data management portal for research projects in the biomedical and life sciences. It is a featured tool from the Tool Pool organised by TMF.
The menoci modules are actively developed and maintained by the University Medical Center Göttingen, Department of Medical Informatics, and Göttingen Scientific Data Center, GWDG.
menoci Source Code
The menoci suite is distributed via the dedicated menoci GitLab repository.
Original Repositories
![]()
This distribution repository comprises the following menoci modules:
- Commons: shared libraries, group and subproject handling
- Data Archive: store and share data packages (using CDSTAR as a backend storage service; https://gitlab.gwdg.de/cdstar/cdstar)
- Literature: publication list
- Wikidata: optional extension of the publication list, that cross-references Wikidata pages of registered articles
- Antibody Catalogue: track antibodies used in the project
- Mouse Line Catalogue: track mouse lines and specimen used in the project
License
Website/Documentation
This documentation is created with mdbook.
The source code of this website is available at menoci-docs GitLab Repository
menoci Project
Copyright (C) 2012-2020 menoci contributors, see AUTHORS.md
The menoci project source code is licensed under GNU General Public License 3.0, see license file
Documentation
This documentation is licensed under Creative Commons license CC-BY-SA 4.0 unless otherwise stated.
Download
Get the latest version from our public distribution GitLab Repository.
Releases
Docker Version
The latest release of menoci is also dockerized and it can be found in the GitLab Registry.
The Docker image with the tag menoci:latest represents the latest release.
Initial Source Code
The following official distribution release versions of the menoci project are available:
| Version | Date | DOI | Comment |
|---|---|---|---|
| 1.0 | 2020-02-05 | doi:10.25625/NC9TF6 | Initial menoci release |
| 1.1 | 2023-03-31 | doi:10.25625/NC9TF6 | Second menoci release |
Installation
On any Linux-based host system with docker and docker-compose packages installed: Get the latest menoci release version, for example by cloning the official Gitlab repository:
git clone https://gitlab.gwdg.de/medinfpub/menoci.gitcd menocito enter the new directory
For a quick exploration, use the docker-compose setup:
- start with command
docker-compose up -d - Open
localhostin web browser, proceed with Drupal installation (choose SQLite database since no other database is available for this container)
More permanent installation should be done via HTTPS and preferably to path /opt/docker/menoci
When using docker-compose.yml file to orchestrate webserver and database
containers, be sure to correctly map the database service name from the compose file
to database hostname during Drupal installation:
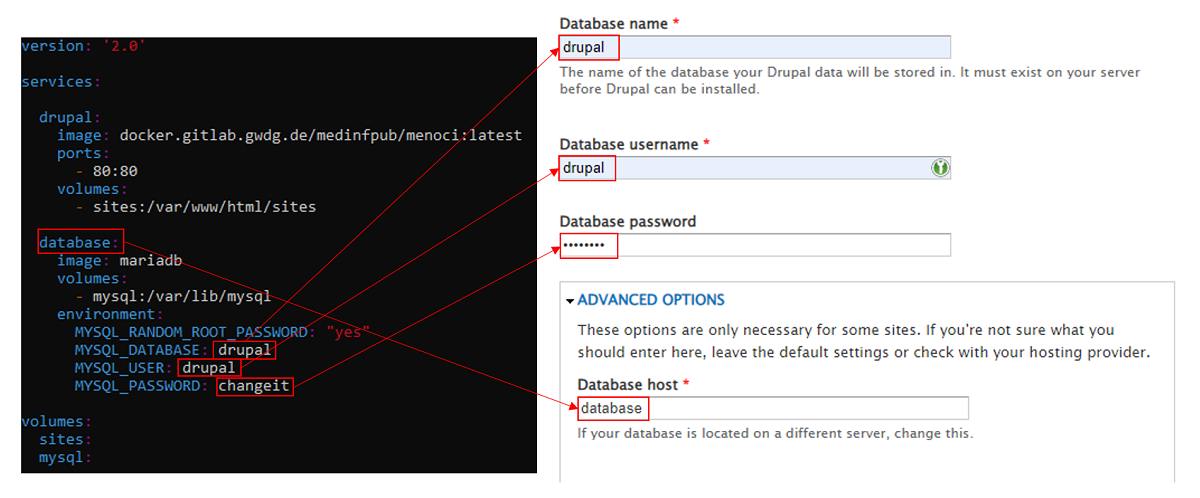
Database choice
- Installation with MySQL/MariaDB as a database server is recommenced.
- SQLite may be a valid choice for testing purposes but some Drupal database mapping may diverge between engines.
- Only MySQL/MariaDB is tested and validated by the development team.
Proxy HTTPS Settings
If you operate the menoci container behind a reverse proxy that adds HTTPS support, additional configuration is required to tell the Drupal instance about its base URL:
- First you need to fetch the
settings.phpfrom the running Drupal/menoci Docker container:
docker cp drupal_container:/var/www/html/sites/default/settings.php /local/path(paths may differ in your installation...) - Edit
settings.php: Set the variable$base_urlto your full URL includinghttps://protocol - Re-transfer the edited file into the container file system:
docker cp /local/path/settings.php drupal_container:/var/www/html/sites/default/settings.php - Restart the container,
docker restart drupal_container - if settings.php is not writable permissions need to be changed
chmod 644 sites/default/settings.php- and
chown www-data: site/default/settings.php
Configuration
After installing Drupal and the menoci modules, some basic settings have to be configured through the Drupal administration user interface.
Configuration instructions on this page are currently work in progress.
Modules
Navigate your browser to the route admin/modules/ of your Drupal instance.
- Enable "jQuery Update" module in the section "User Interface"
Appearance
Go to admin/appearance/
-
Enable "Bootstrap" theme and set as default.
-
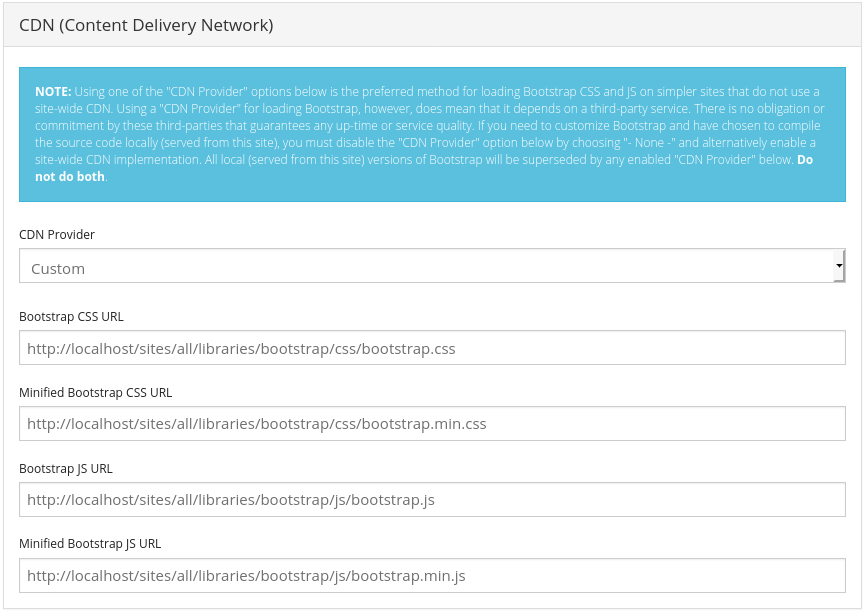
[optional] In Bootstrap settings, "Advanced": set "CDN provider" to "custom" and replace default paths (like
https://cdn.jsdelivr.net/npm/bootstrap@3.4.1/dist/*) with
/sites/all/libraries/bootstrap. The result should look like this for the the first CSS parameter:
/sites/all/libraries/bootstrap/css/bootstrap.css
respectively for the CSS and JS fields. (This is optional but Bootstrap library files are shipped with the default Docker container, allowing you to minimize CDN traffic and related user tracking.)
Modules
Go to admin/modules/
- Enable "Commons" module, press "save" button, and confirm that module dependencies will be automatically installed.
- Enable "Published Data Registry" module in the section "Menoci.io".
- Choose from the following optional menoci modules to install:
- Mouseline Catalogue
- Research Data Archive
- Wikidata extension
Configuration
Go to admin/config/
-
within the section
Search and metadataenableclean URLs -
Create first research group
-
[optional] Create subprojects
Development
Development of menoci is actively pursued at the Göttingen Department of Medical Informatics. Please find below a rough outline of release versions and latest released features.
The menoci project is subject to change. If you would like to see certain features implemented on schedule, feel free to contact us and contribute to development.
WIP
- API and swagger documentation: To extend the accessibility of research data stored in menoci the Swagger-UI documentation for the menoci API has been added to the project.
- Demo instance: menoci and can be tried out and tested with the demo instance. This instance is free to use and publicly available and will be resetted each night.
Privacy policy
The menoci.io web site is run by the University Medical Center Göttingen, Department of Medical Infomatics.
When this policy refers to “we”, this means the above Department of Medical Informatics.
We strictly limit the processing of your personal information, and in general process as little data as possible to fulfill our mission. We won't sell your data to any third party.
Whom to contact
If you have questions please contact mi@med.uni-goettingen.de.
This privacy policy has been adapted from the FSFE website
Imprint
University Medical Center Göttingen
Department of Medical Informatics
Prof. Dr. Dagmar Krefting
Head of Department
Robert-Koch-Str. 40
37075 Göttingen, Germany
E-Mail: menoci@medizin.uni-goettingen.de
Phone: +49 551 39-3431